Group8_17
Chih-Hsuan Wu
Last updated: 2024-10-22
Checks: 7 0
Knit directory: DEanalysis/
This reproducible R Markdown analysis was created with workflowr (version 1.7.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230508) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version 35f298a. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.Rhistory
Untracked files:
Untracked: .DS_Store
Untracked: .gitignore
Untracked: analysis/analysis_humanspine.Rmd
Untracked: analysis/simulation_donor_effect.Rmd
Untracked: data/.DS_Store
Untracked: data/10X_inputdata.RData
Untracked: data/10X_inputdata_cpm.RData
Untracked: data/10X_inputdata_integrated.RData
Untracked: data/10X_inputdata_lognorm.RData
Untracked: data/10Xdata_annotate.rds
Untracked: data/Bcells.Rmd
Untracked: data/Bcellsce.rds
Untracked: data/Kang_DEresult.RData
Untracked: data/Kang_data.RData
Untracked: data/fallopian_DEresult.RData
Untracked: data/fallopian_tubes.RData
Untracked: data/fallopian_tubes_all.RData
Untracked: data/human/
Untracked: data/human_spine.RData
Untracked: data/human_spine_DEresult.RData
Untracked: data/ls_offset_Result.RData
Untracked: data/mouse/
Untracked: data/permutation.RData
Untracked: data/permutation13.RData
Untracked: data/permutation2.RData
Untracked: data/splatter_simulation.RData
Untracked: data/vstcounts.Rdata
Untracked: figure/
Unstaged changes:
Modified: analysis/analysis on Kang.Rmd
Modified: analysis/analysis on fallopian tubes.Rmd
Modified: analysis/preprocess_fallopian_tubes.Rmd
Modified: code/DE_methods.R
Modified: code/functions_in_rmd.R
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/group8_17.Rmd) and HTML
(docs/group8_17.html) files. If you’ve configured a remote
Git repository (see ?wflow_git_remote), click on the
hyperlinks in the table below to view the files as they were in that
past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 35f298a | C-HW | 2024-10-22 | same criteria, FCcutoff 1.5 |
| html | 28b9b7d | C-HW | 2024-10-14 | Build site. |
| Rmd | c4961dc | C-HW | 2024-10-14 | update Seurat result |
| html | 5e86686 | C-HW | 2023-12-15 | Build site. |
| Rmd | 036d5fc | C-HW | 2023-12-15 | wflow_publish(c("analysis/group12_13.Rmd", "analysis/group12_19.Rmd", |
| html | d7a7b73 | C-HW | 2023-12-10 | Build site. |
| Rmd | ae4b56c | C-HW | 2023-12-10 | adjust ylim of p-value histogram |
| html | ac027b0 | C-HW | 2023-12-09 | Build site. |
| Rmd | 582be29 | C-HW | 2023-12-09 | update new DE results |
| html | 2a17159 | C-HW | 2023-12-05 | Build site. |
| Rmd | e7f3de4 | C-HW | 2023-12-05 | fix Wilcox log2FC sign |
| html | bc13544 | C-HW | 2023-12-04 | Build site. |
| Rmd | 39196a3 | C-HW | 2023-12-04 | fix log2FC sign |
| html | 42900f0 | C-HW | 2023-12-01 | Build site. |
| Rmd | c774a2d | C-HW | 2023-12-01 | modify method title |
| Rmd | 031d955 | C-HW | 2023-12-01 | create t score comparison |
| html | 031d955 | C-HW | 2023-12-01 | create t score comparison |
| Rmd | 3803697 | C-HW | 2023-12-01 | upload rmd |
| html | 59b08c2 | C-HW | 2023-11-29 | update index, FD permuation, plots axes |
| html | e8b0519 | C-HW | 2023-11-29 | update all pairs |
| html | d7d838c | C-HW | 2023-08-11 | update graph |
| html | ac090b4 | C-HW | 2023-08-03 | update 8_17 heatmap |
| html | 0e93efd | C-HW | 2023-07-26 | upload again |
| html | f314434 | C-HW | 2023-07-26 | add muscat methods |
| html | 7ee9782 | C-HW | 2023-07-13 | add 8_17 |
Data summary
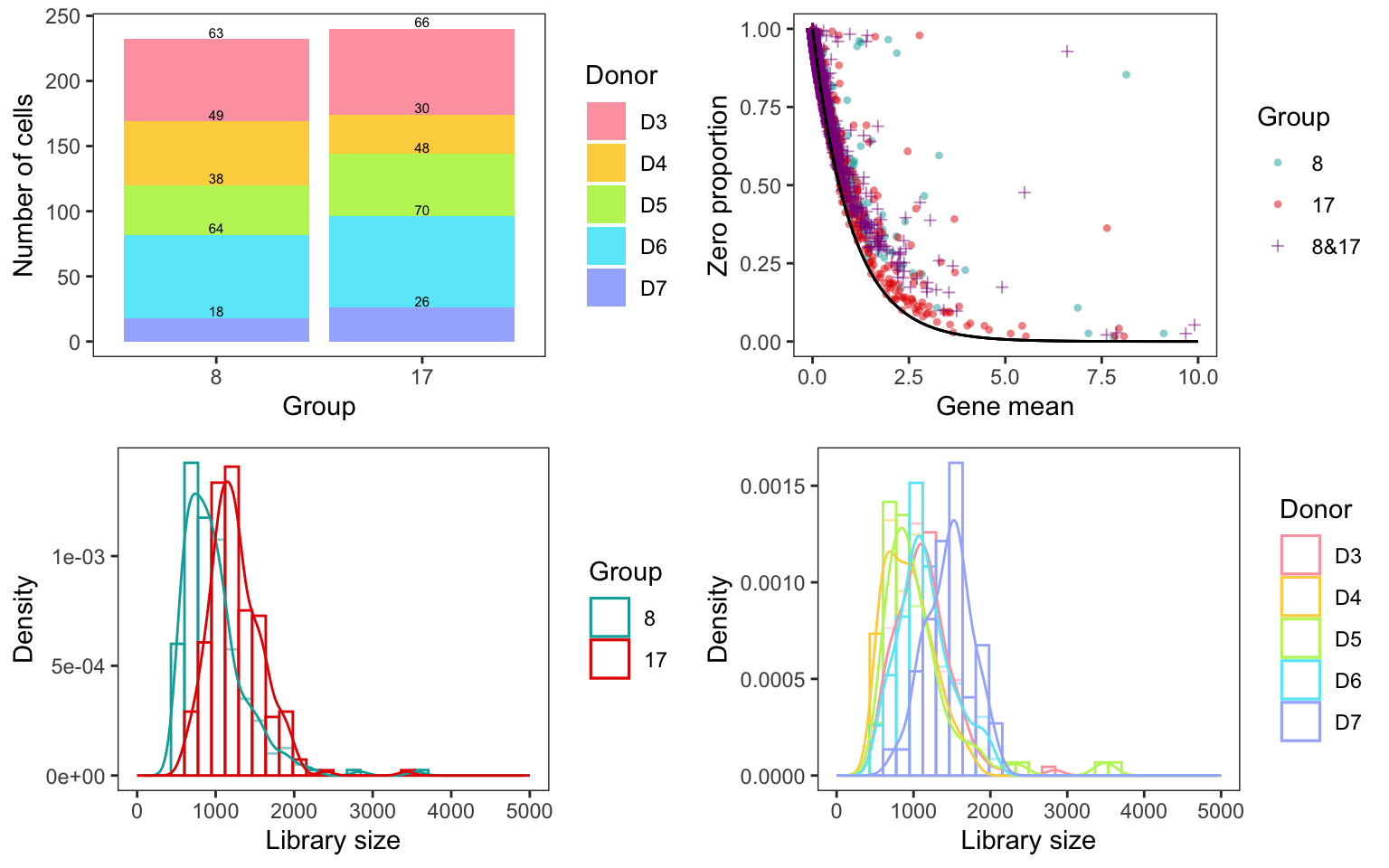
Difference in library size
Asymptotic two-sample Kolmogorov-Smirnov test
data: subset(count_cell_df, Group == group1, totalcount)$totalcount and subset(count_cell_df, Group == group2, totalcount)$totalcount
D = 0.44612, p-value < 2.2e-16
alternative hypothesis: two-sided
Welch Two Sample t-test
data: subset(count_cell_df, Group == group1, totalcount)$totalcount and subset(count_cell_df, Group == group2, totalcount)$totalcount
t = -5.1593, df = 333.02, p-value = 4.267e-07
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-456.1859 -204.3422
sample estimates:
mean of x mean of y
982.0776 1312.3417 Mean difference in raw data/normalized data
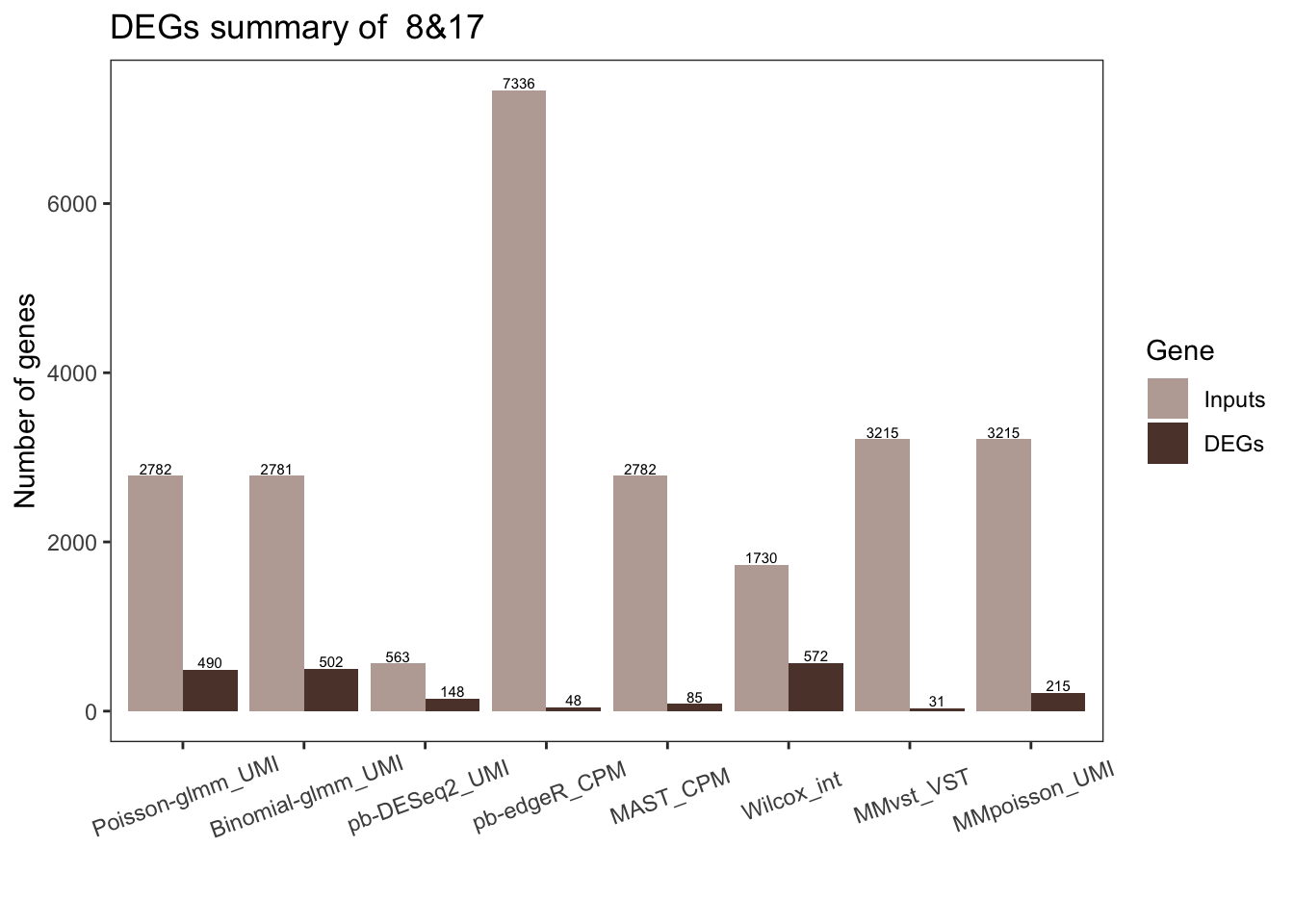
Number of DEGs from each method
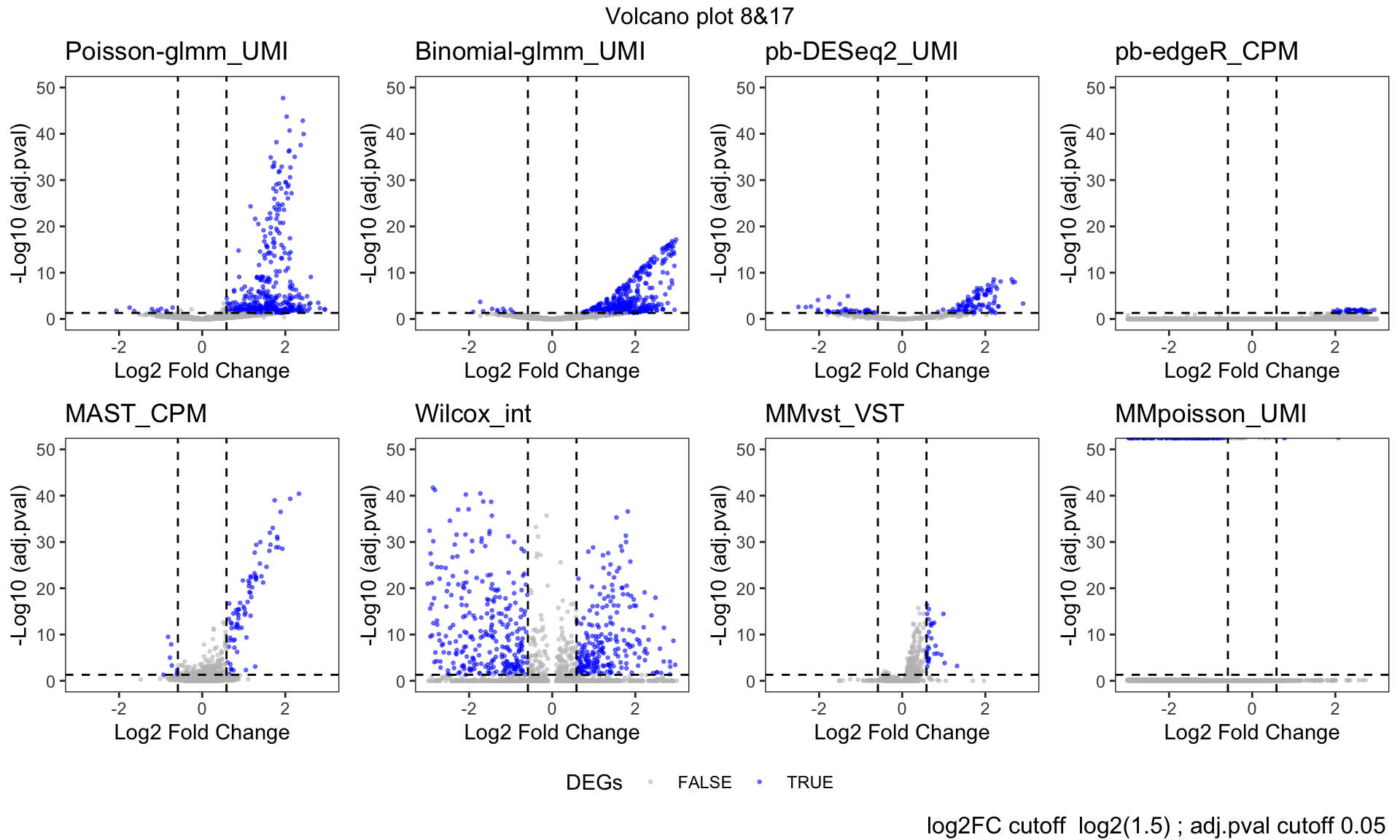
Volcano plot
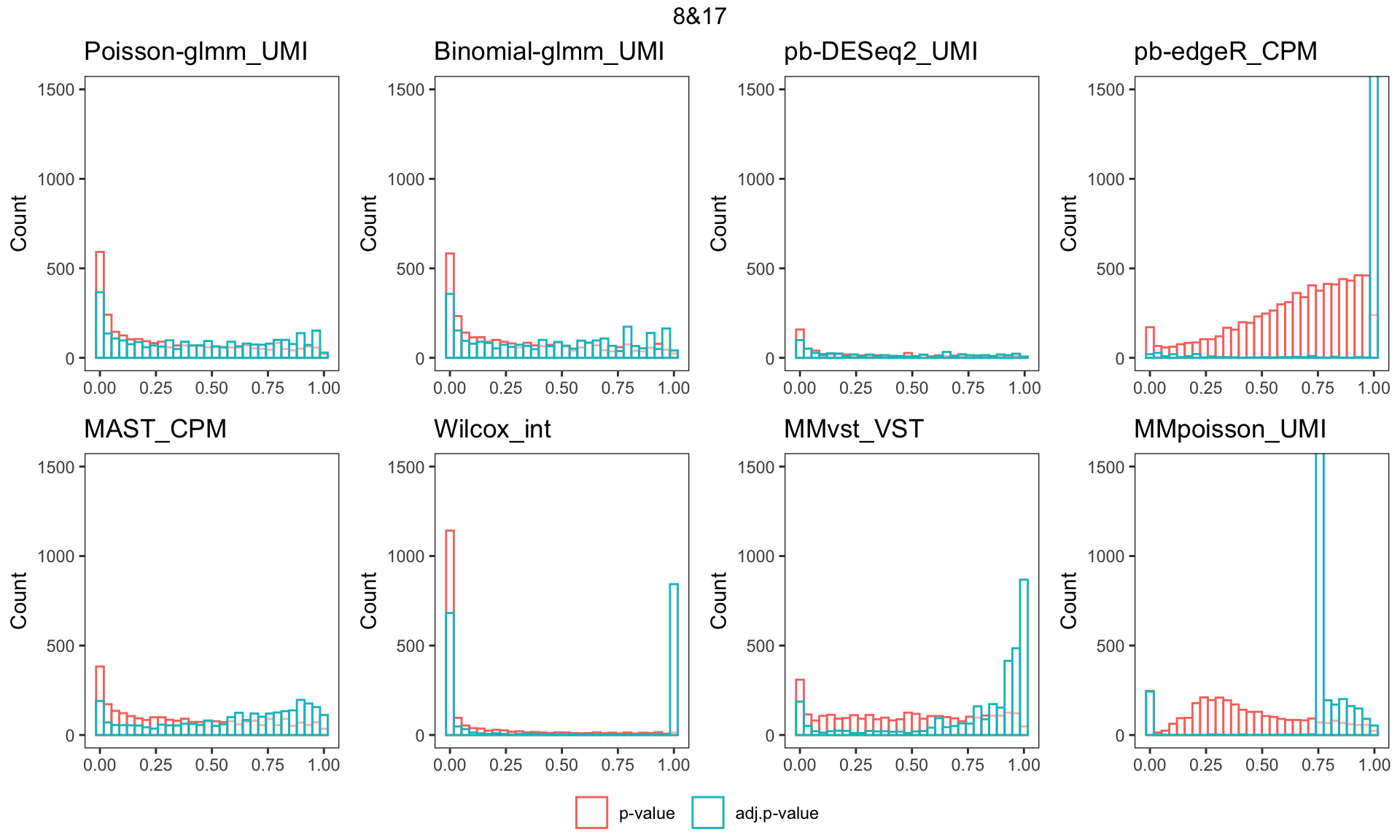
Histogram of p-value/adj.p-value
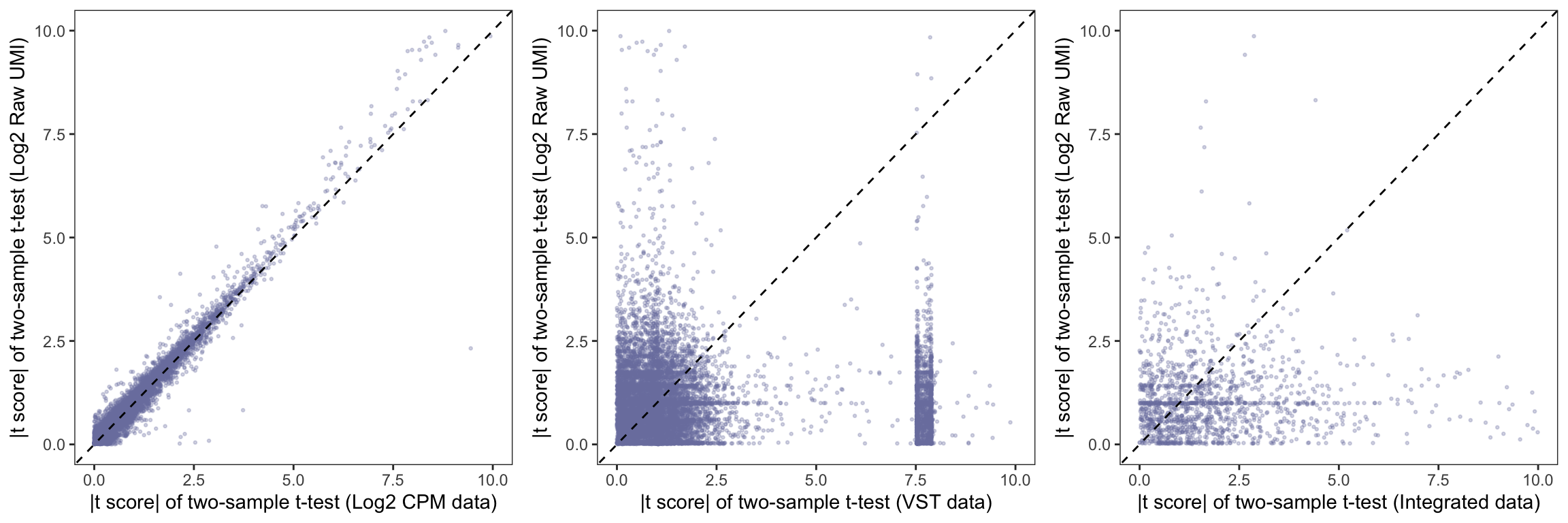
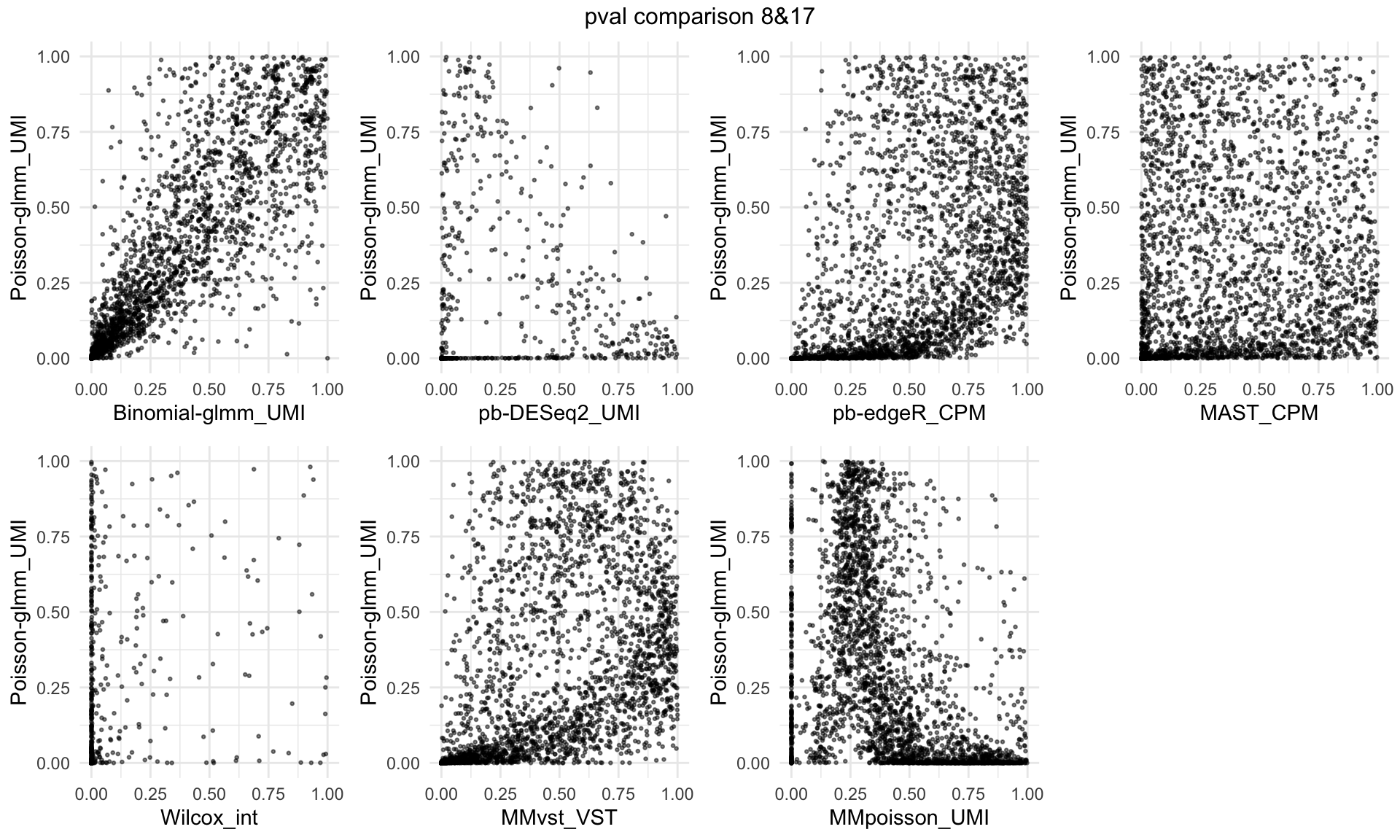
P-Value comparison across different methods
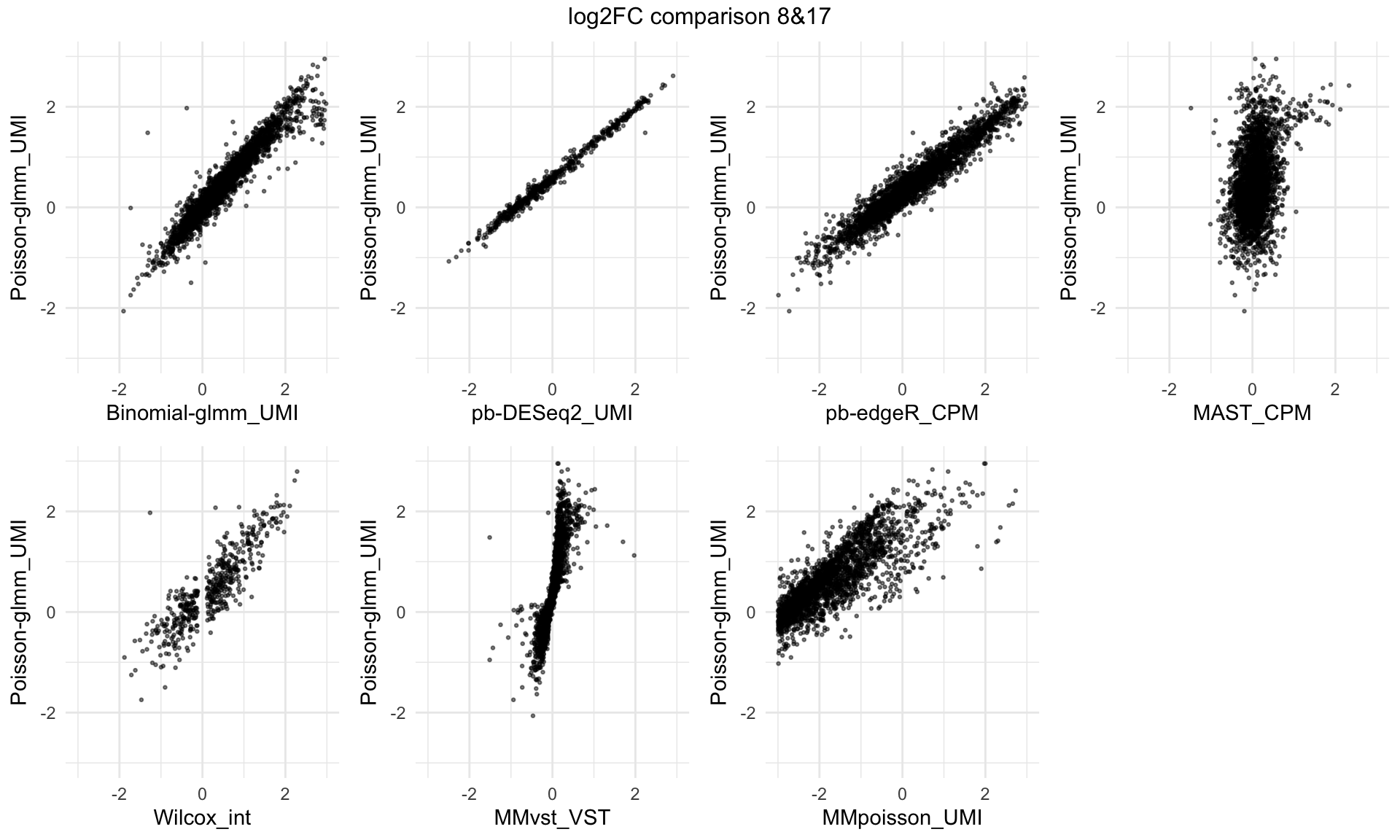
Log2 fold change comparison across different methods
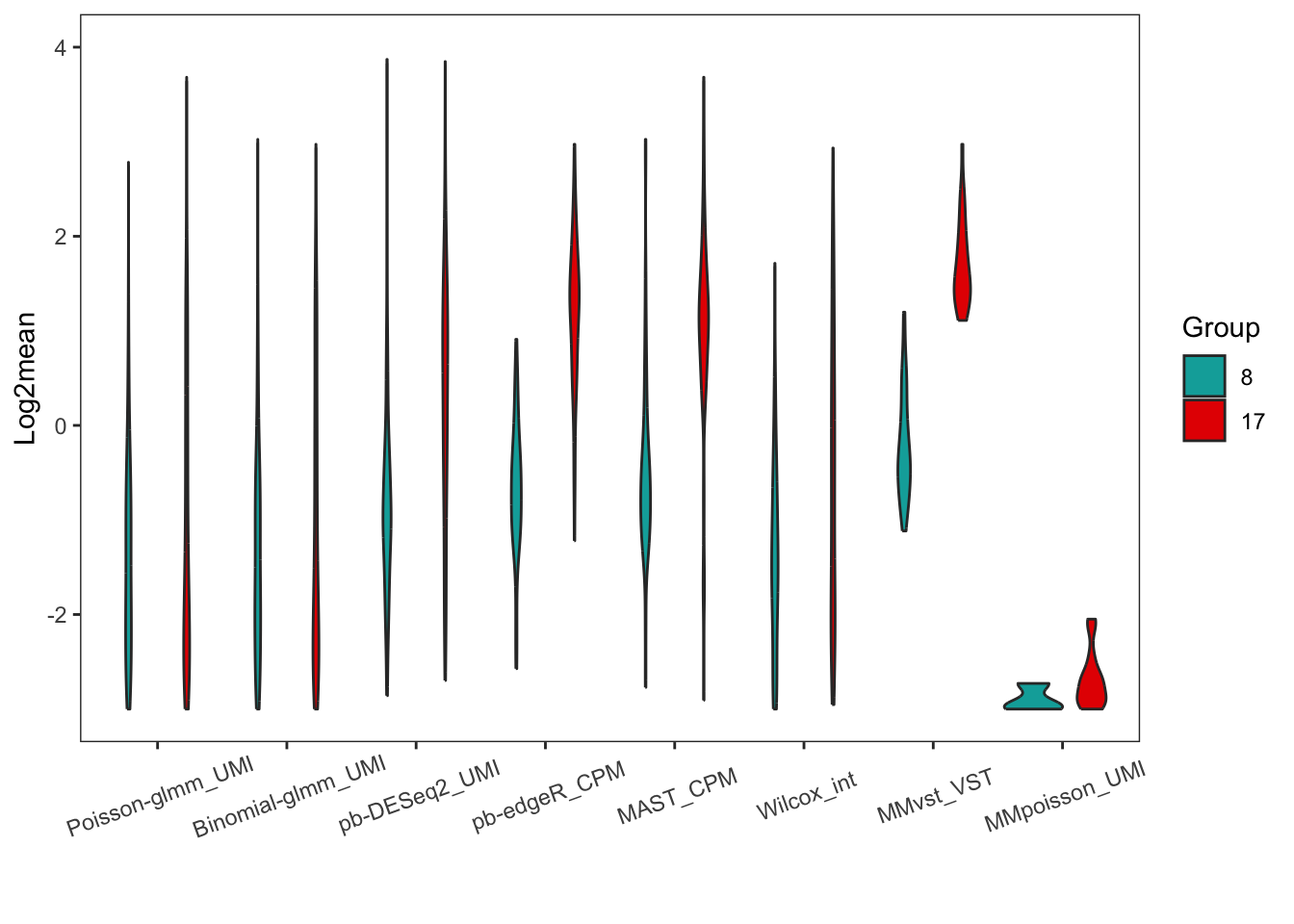
Violin plot of log2mean of DEGs
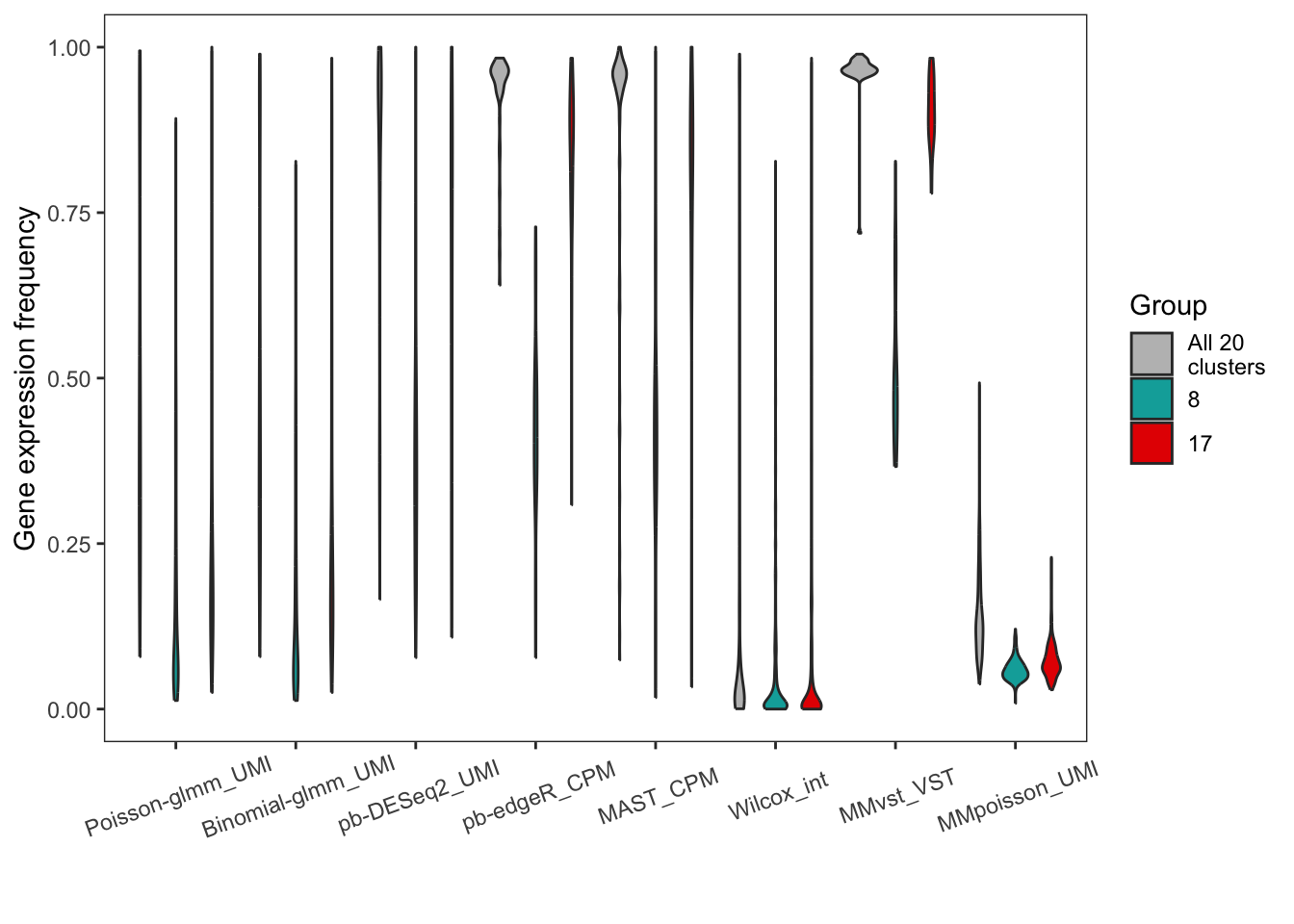
Violin plot of gene expression frequency of DEGs
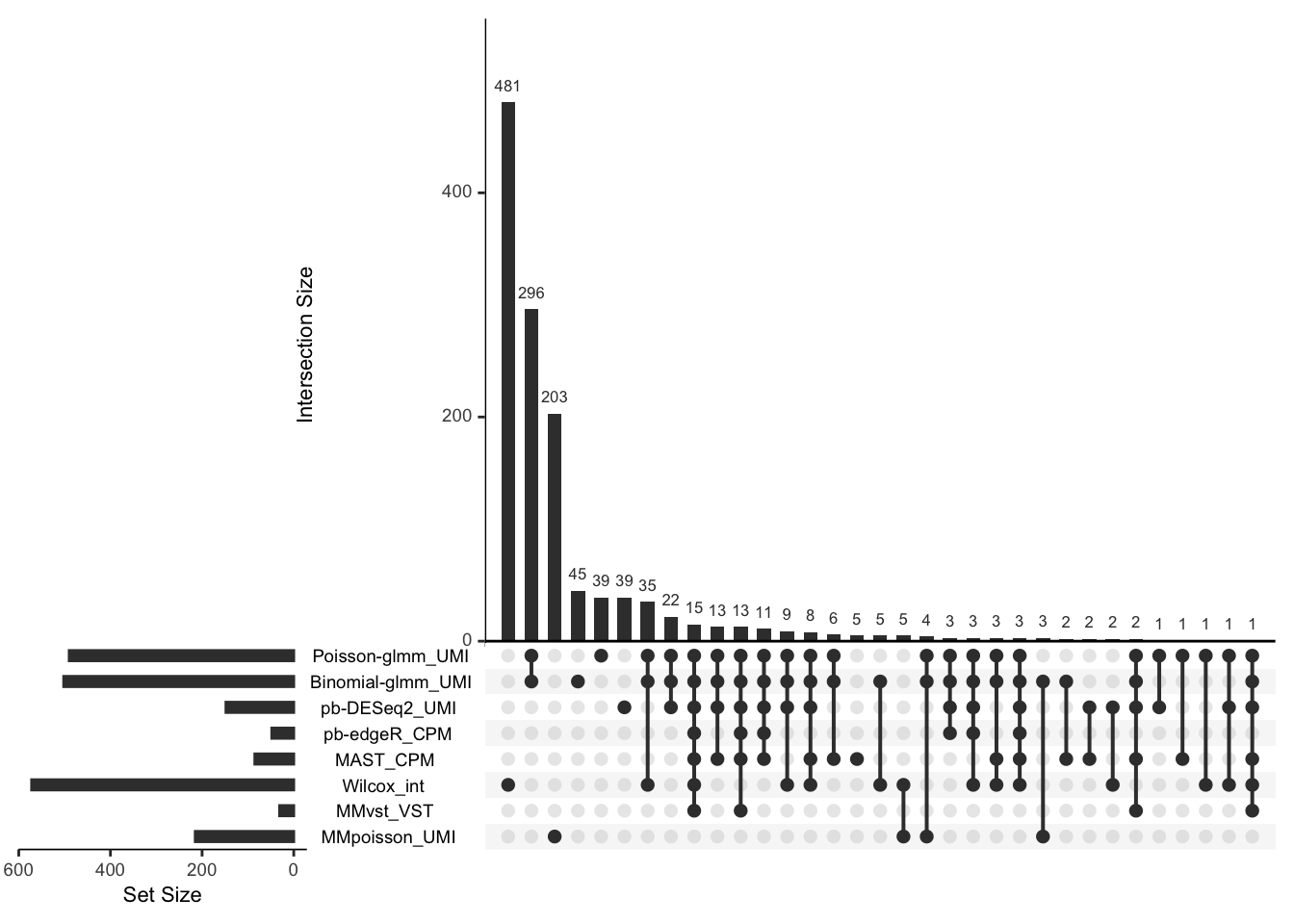
Upset plot
Heatmap of top DEGs
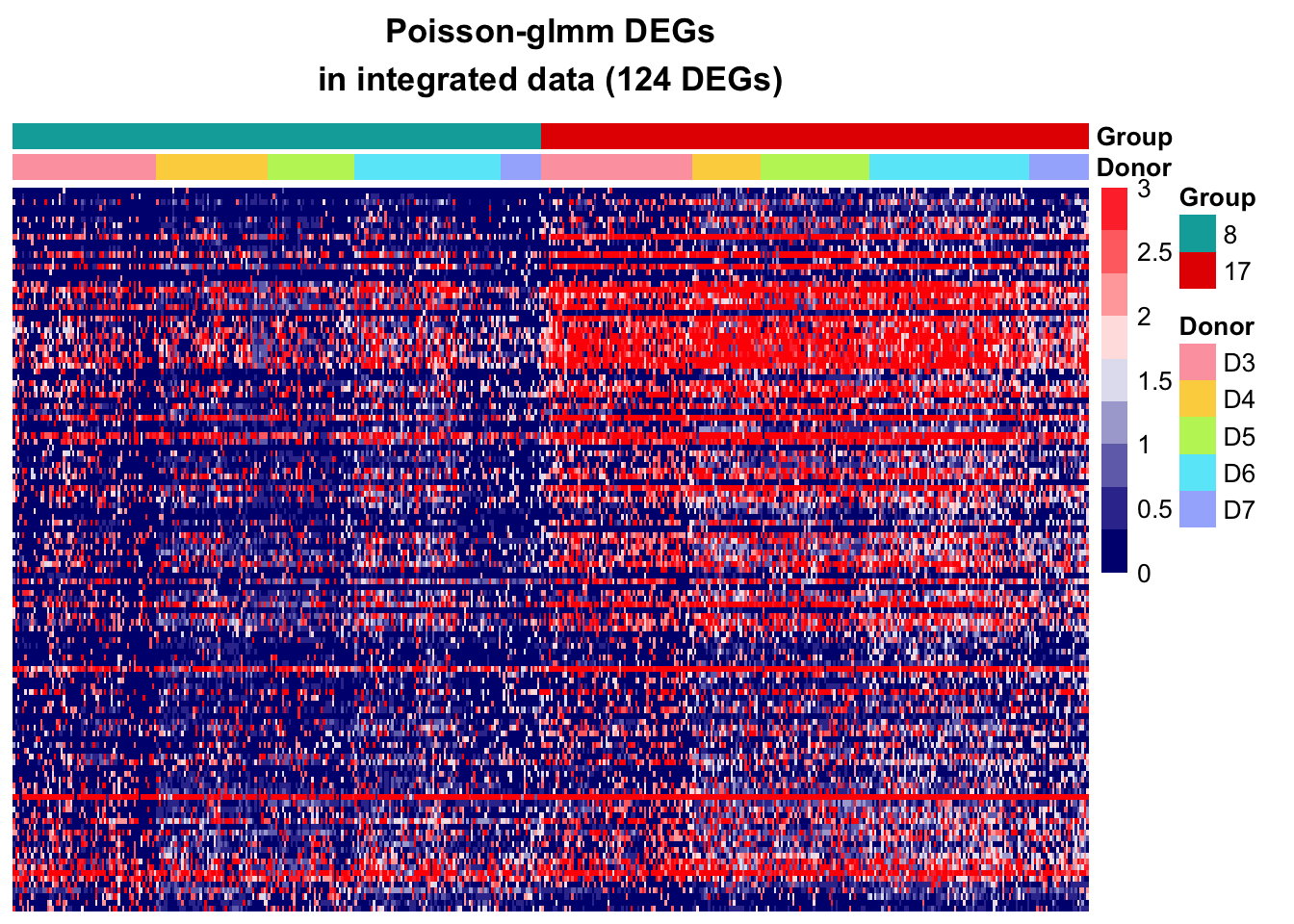
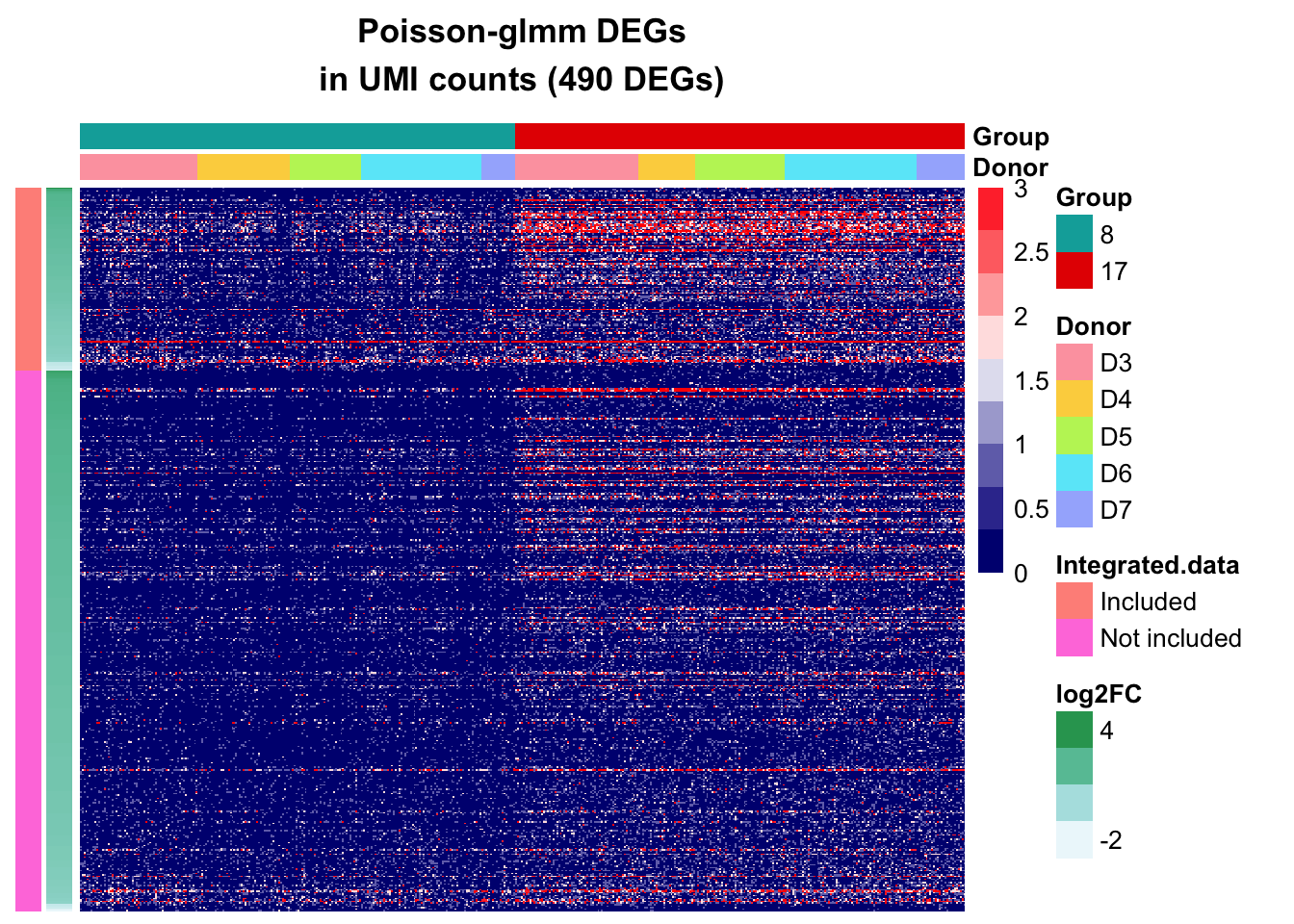
Poisson-glmm DEGs
UMI counts
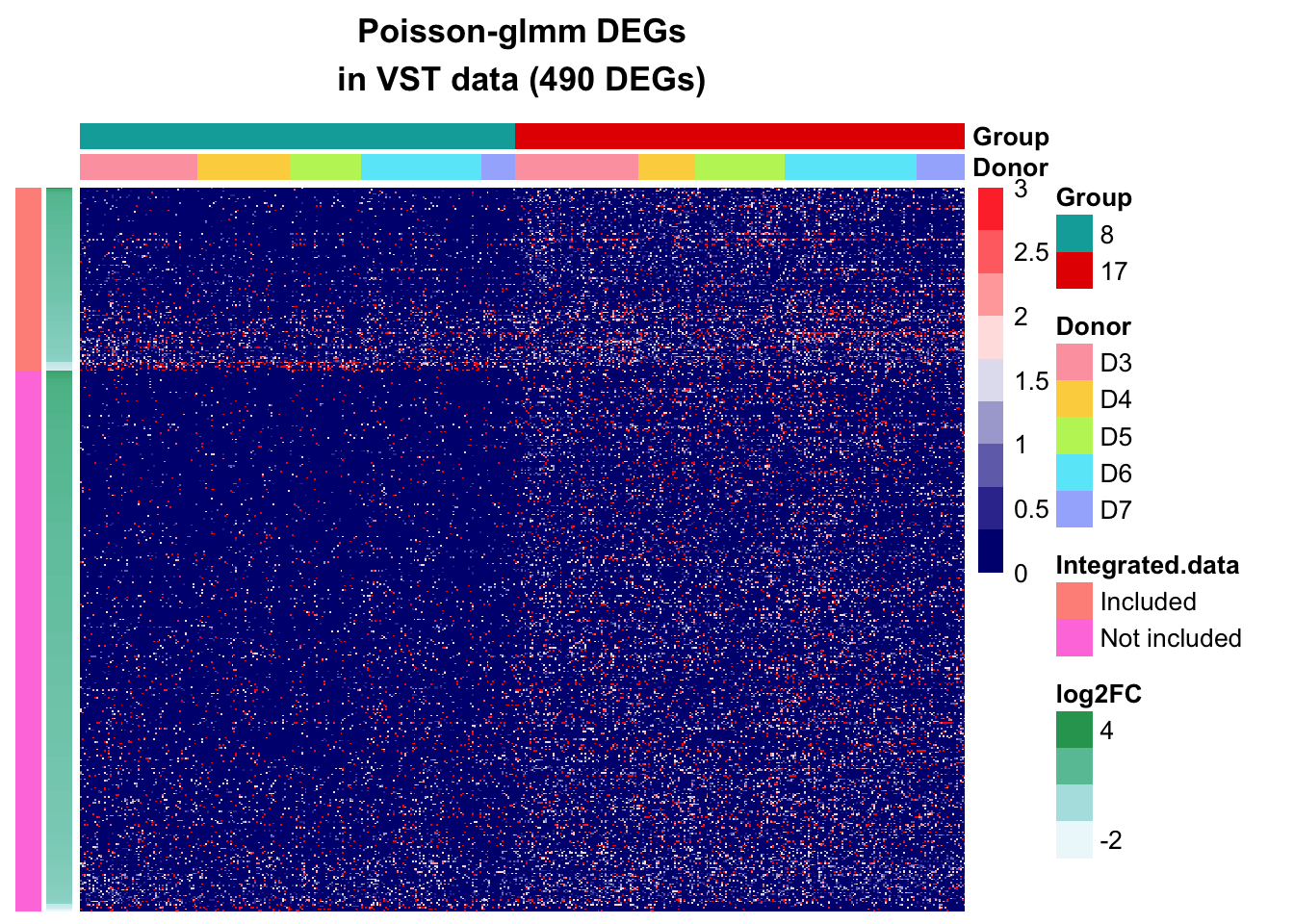
VST data
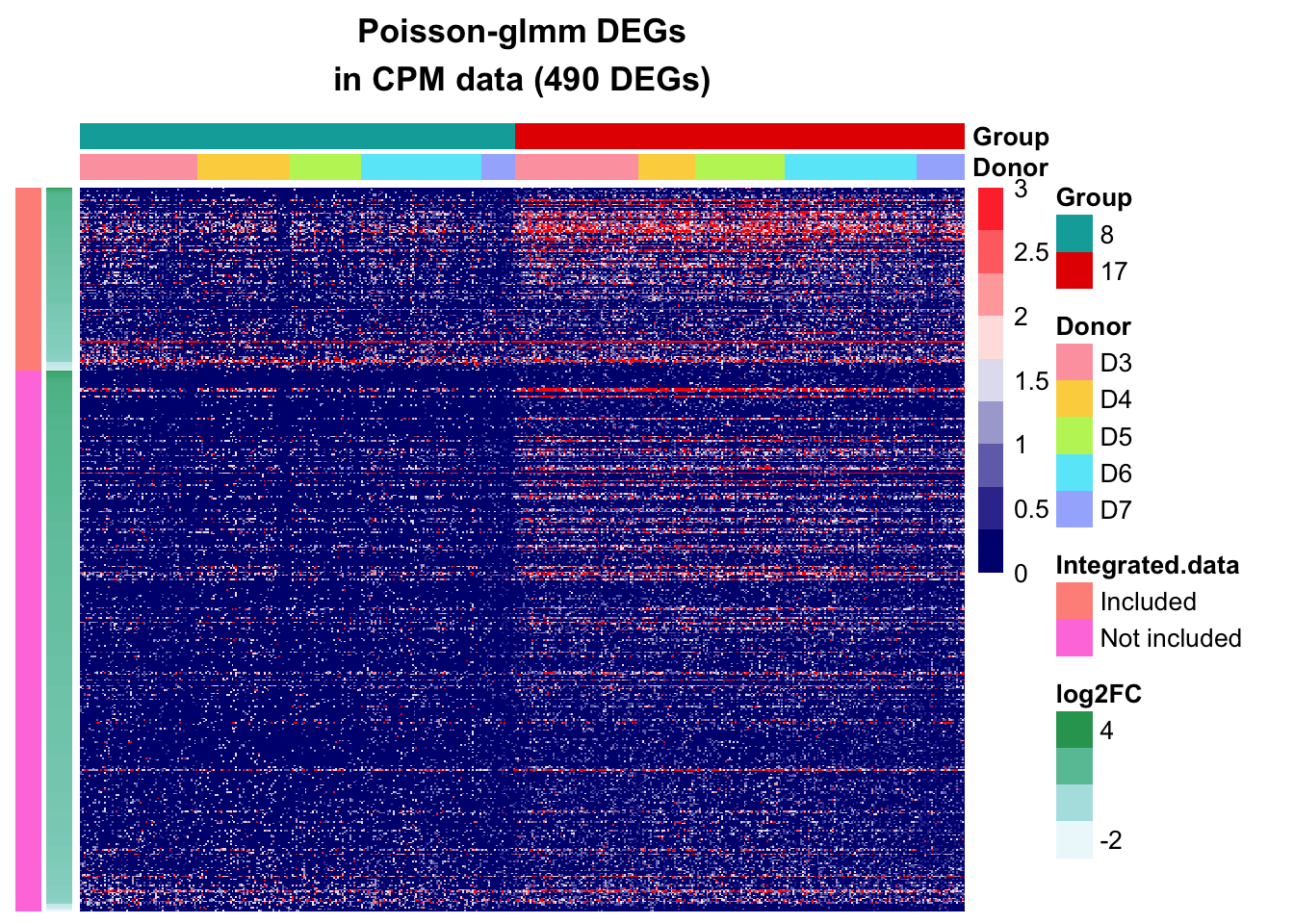
CPM data
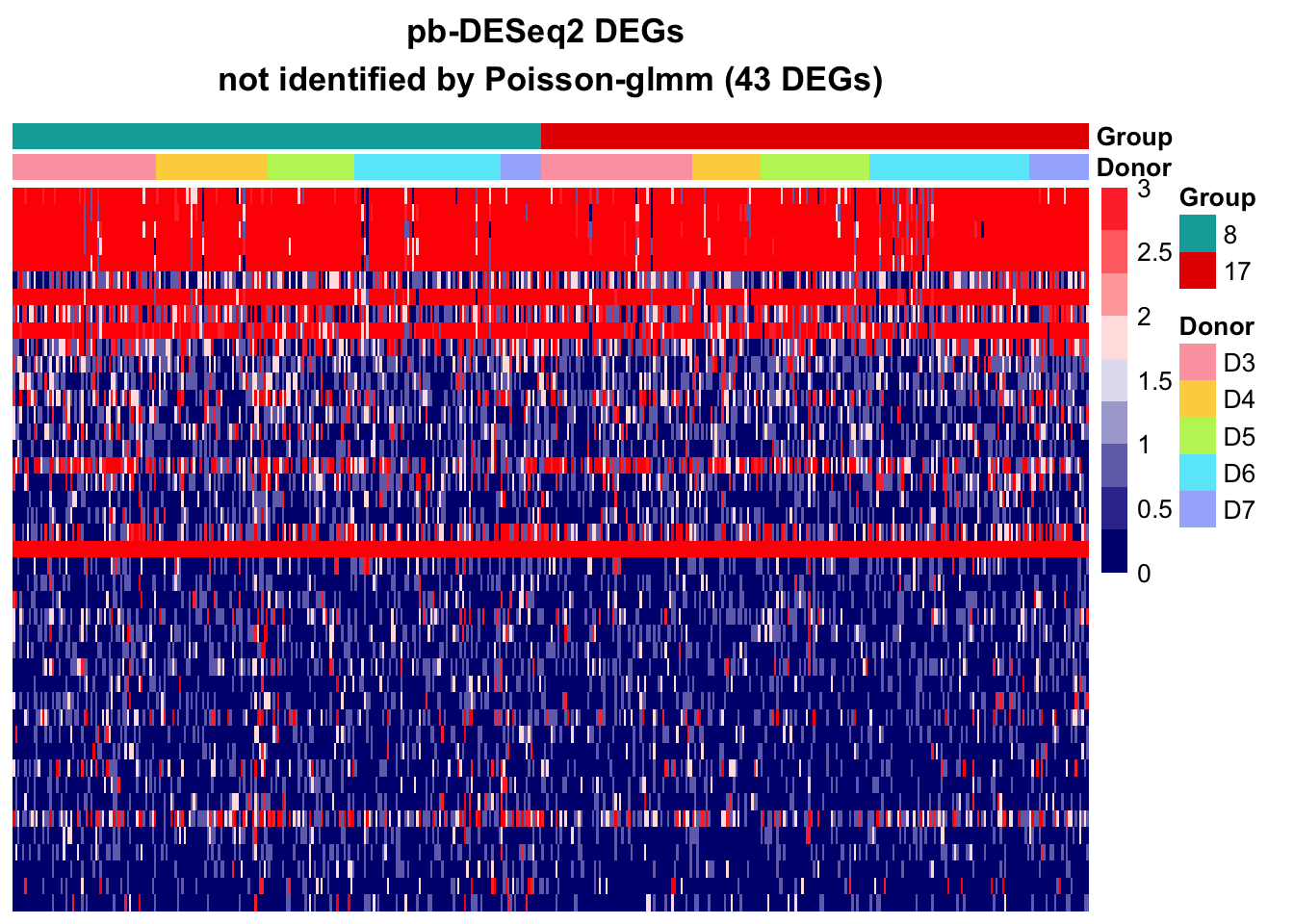
Additional DEGs from other methods
pb-DESeq2
Binomial-glmm
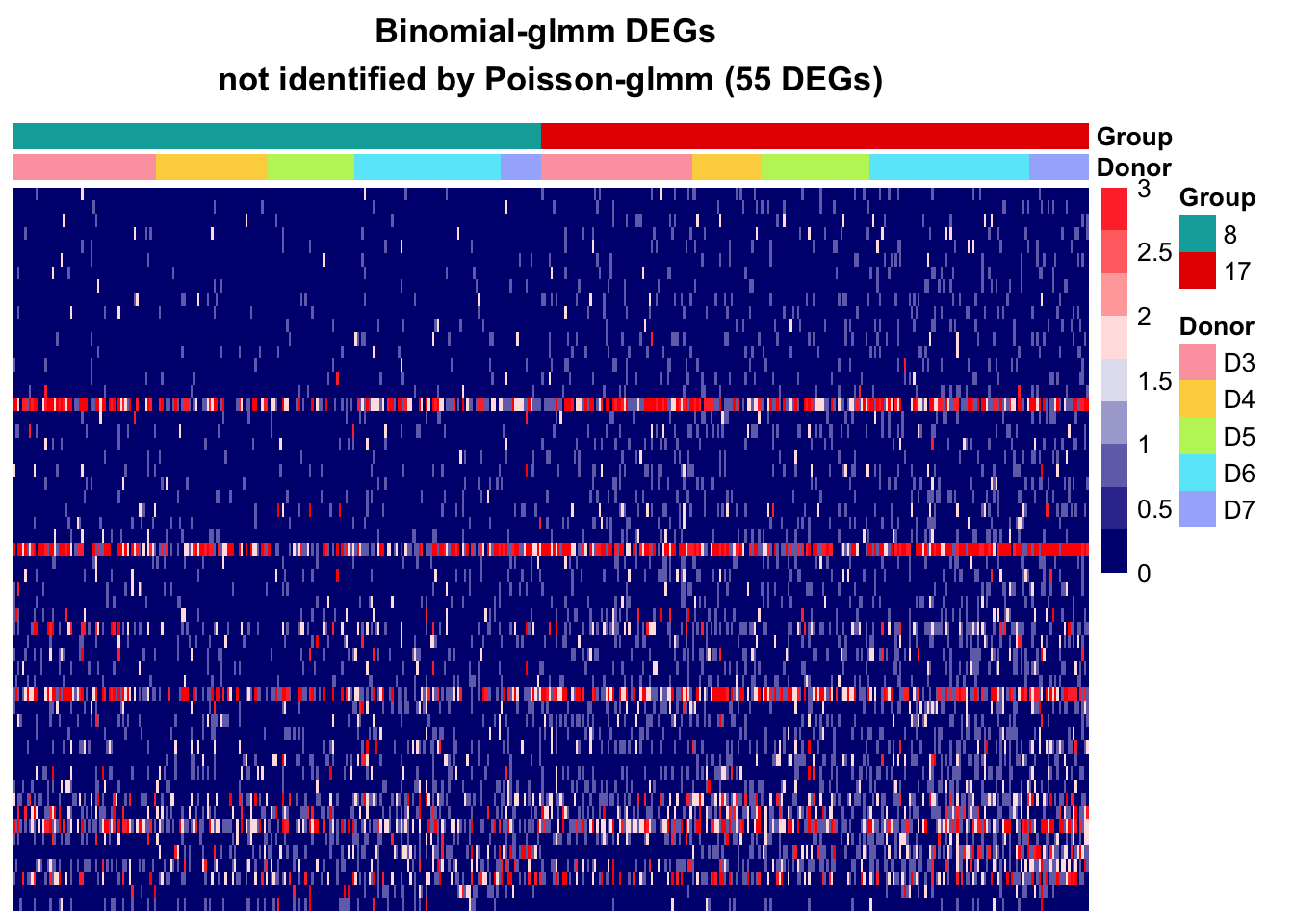
Wilcox
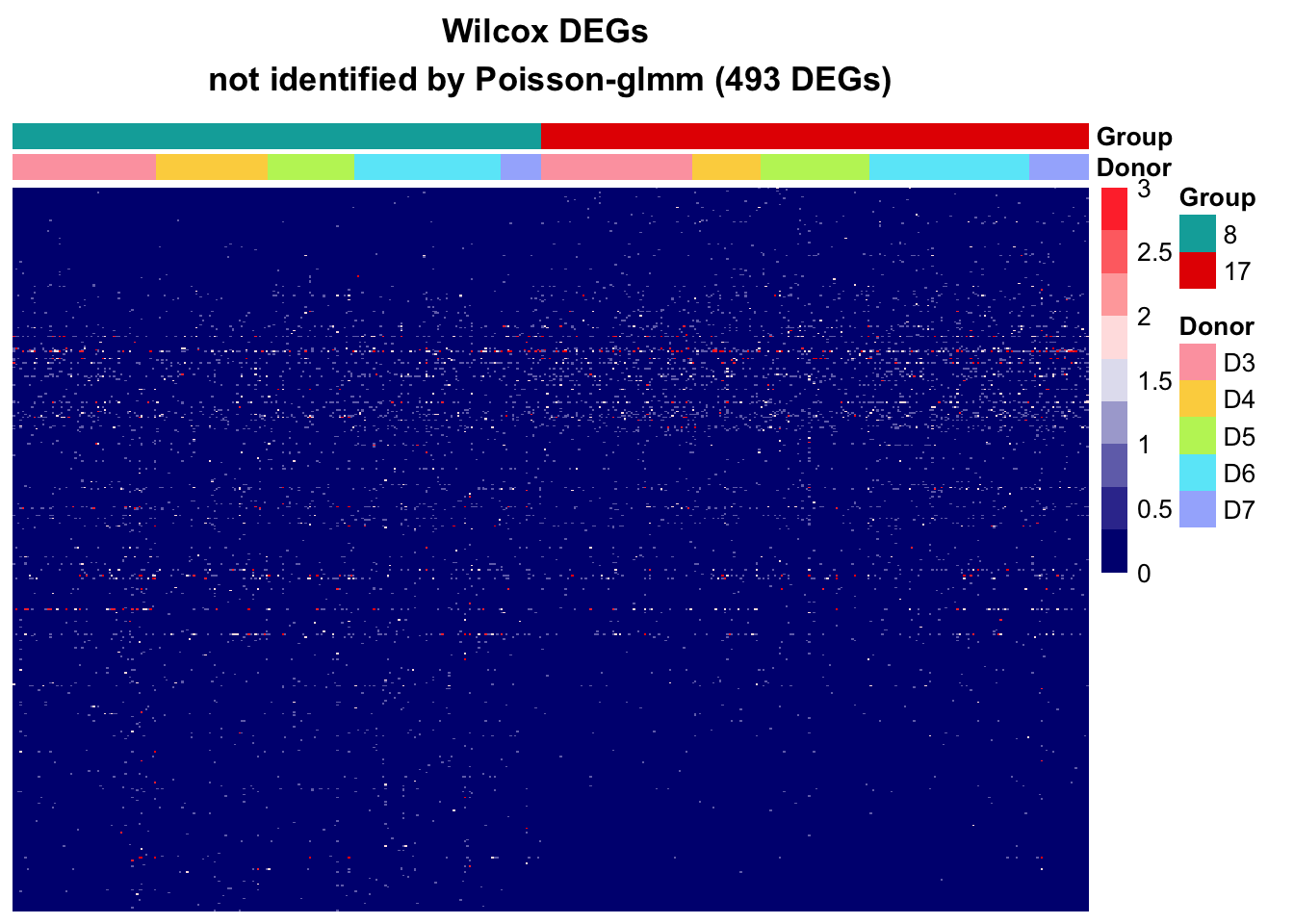
MAST
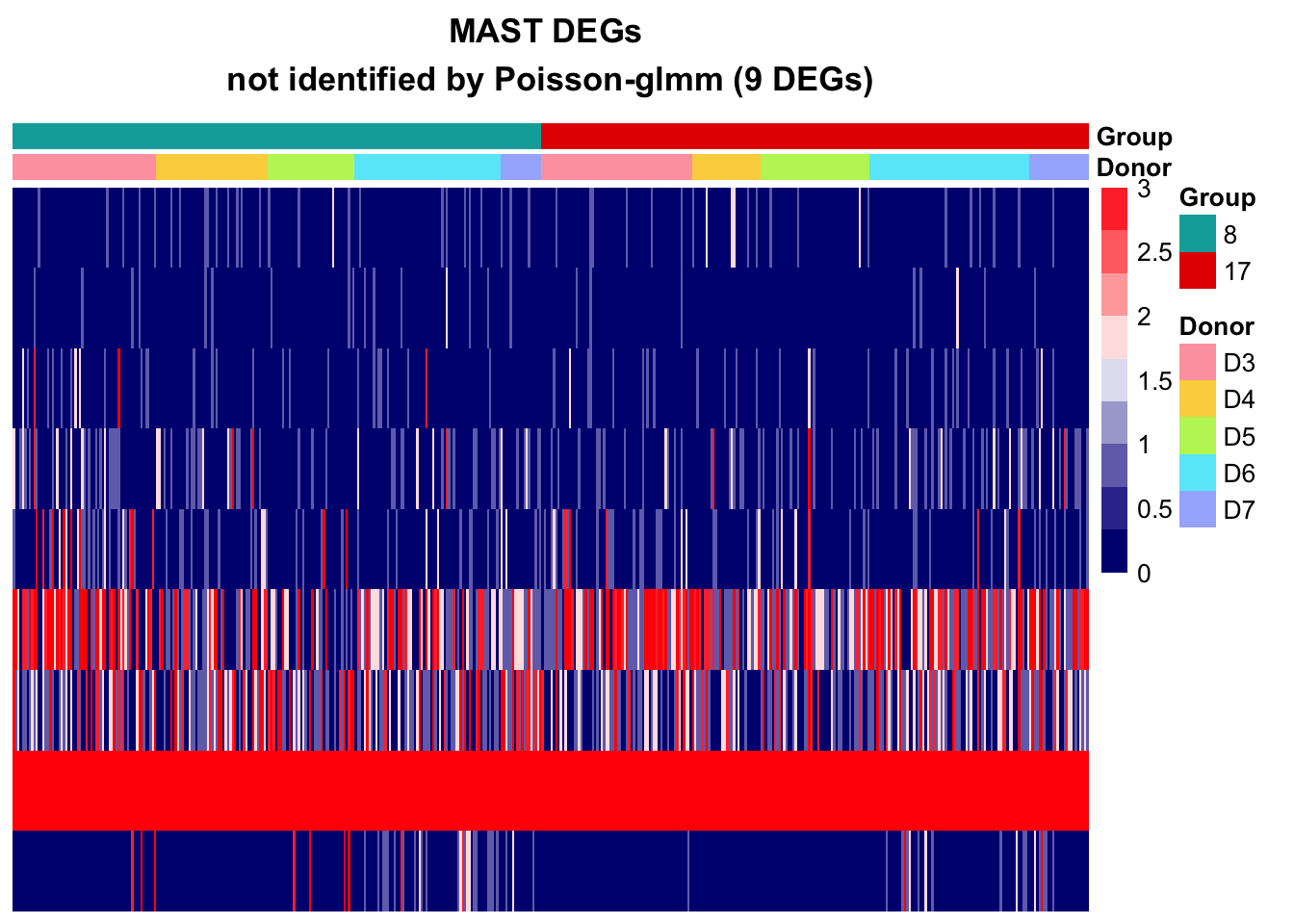
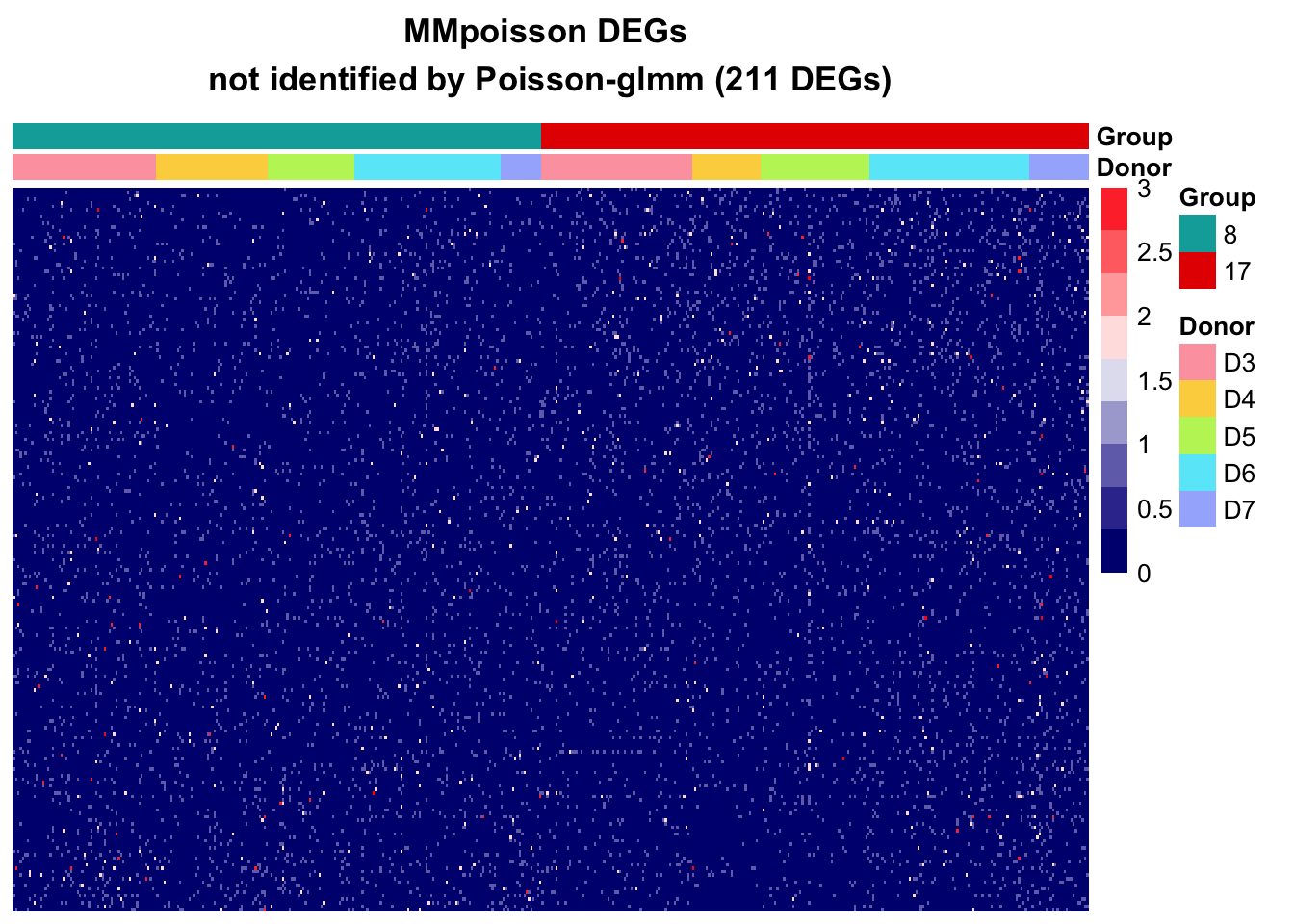
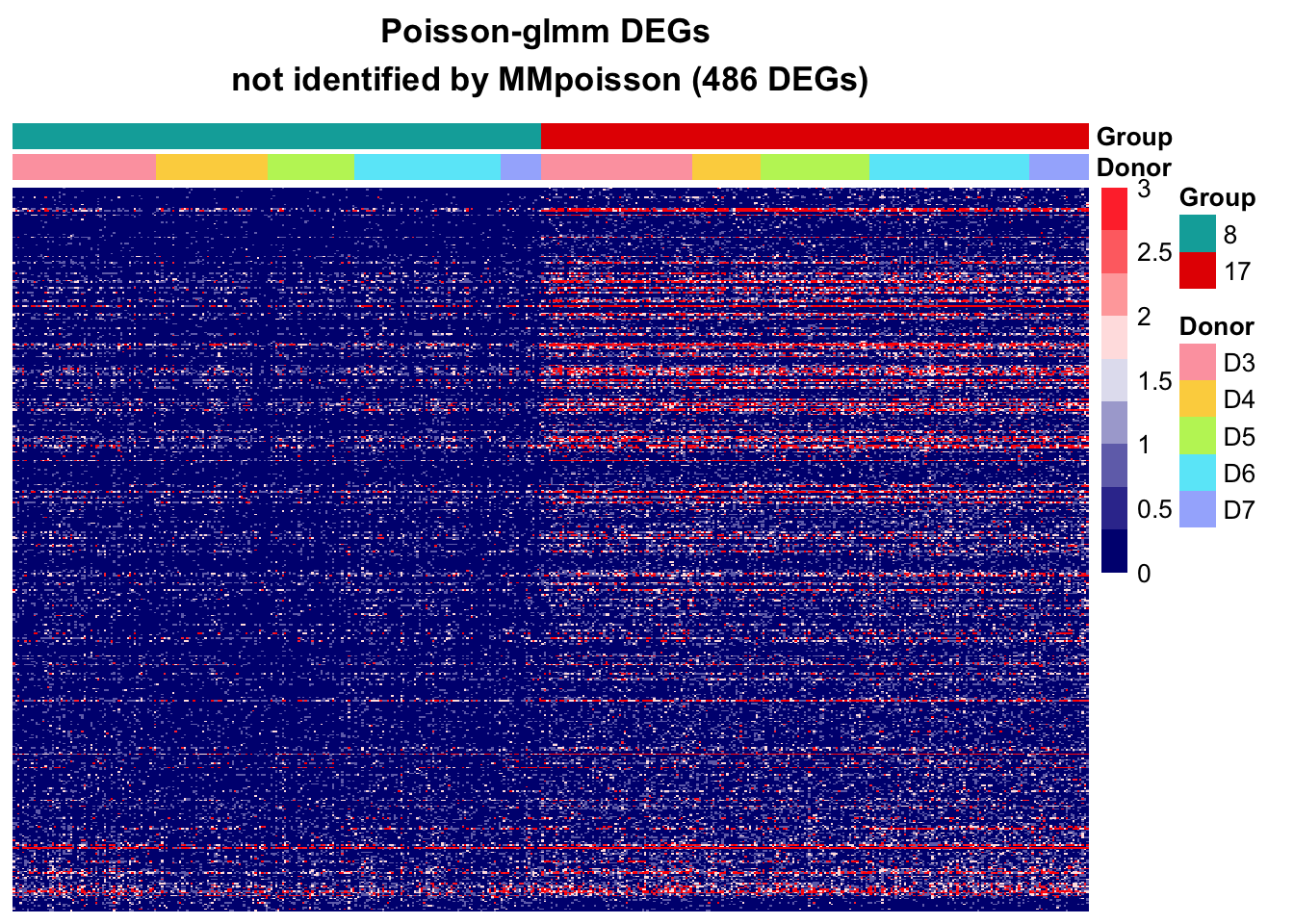
DEGs in Poisson-glmm not identified by MMpoisson
In the MMpoisson model, cell type is considered as a random effect. This approach treats certain aspects of cell type variations as random factors. Consequently, it may obscure the true variation in cell types, limiting its ability to accurately reveal the specific differences between different cell types.
Additionally, the library size is employed as an offset to normalize the counts. That is, the model is considering rate instead of counts. Suppose some genes are highly expressed in one cell type than the other, the absolute difference could be eliminate after accounting for library size. This normalization approach may inadvertently mask certain gene expression differences between cell types.
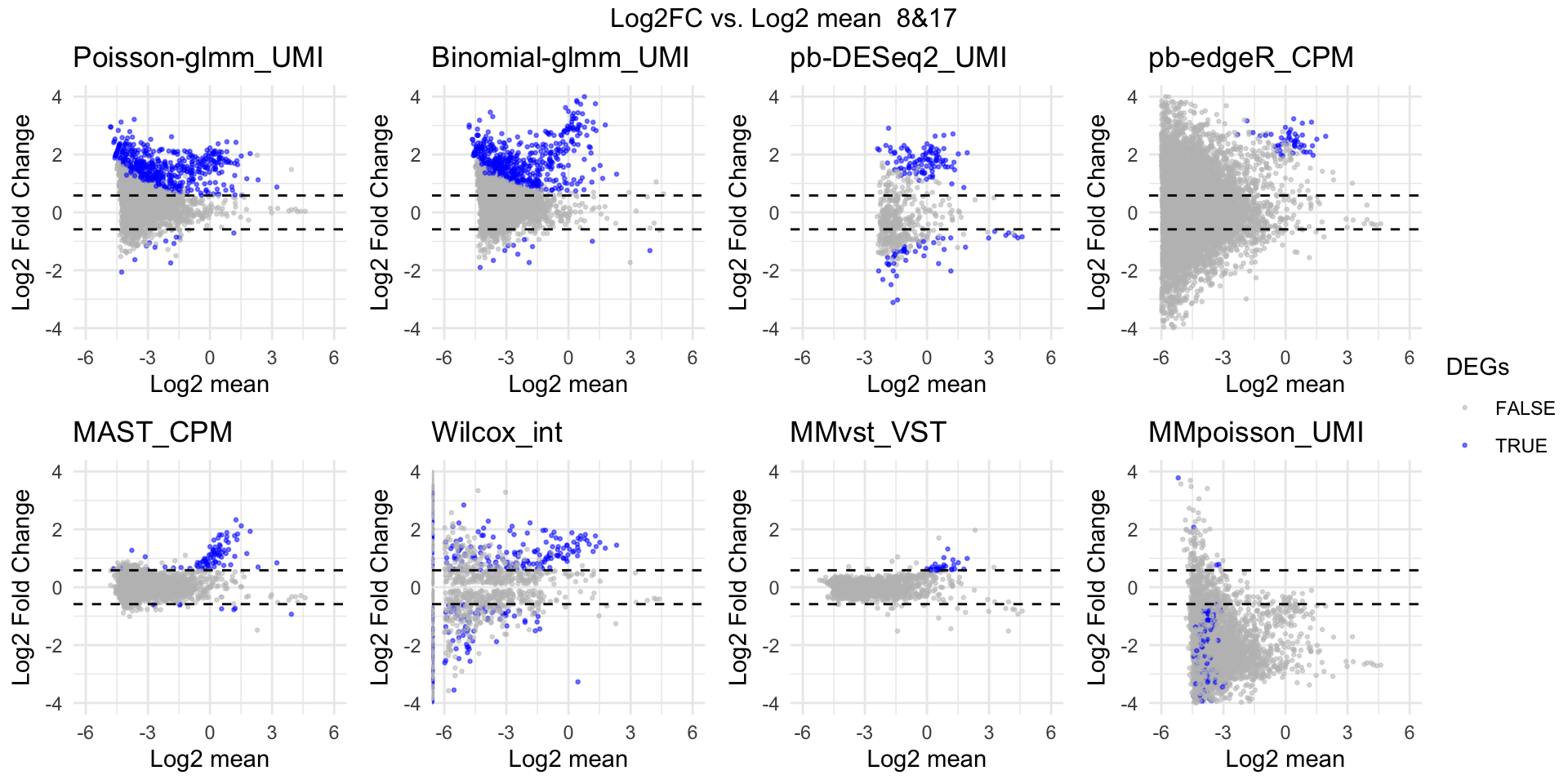
MA plot
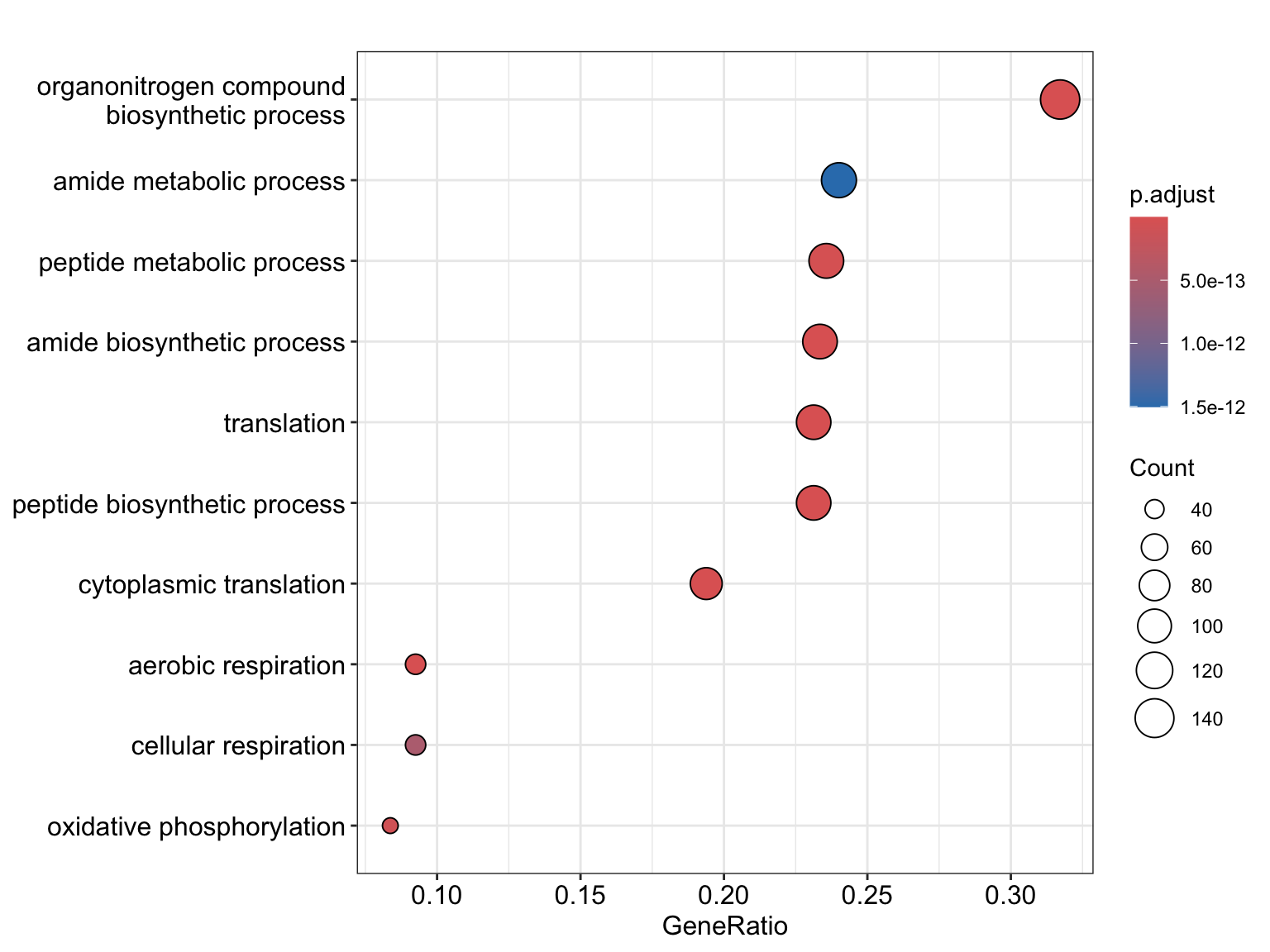
Enrichment analysis
GO object

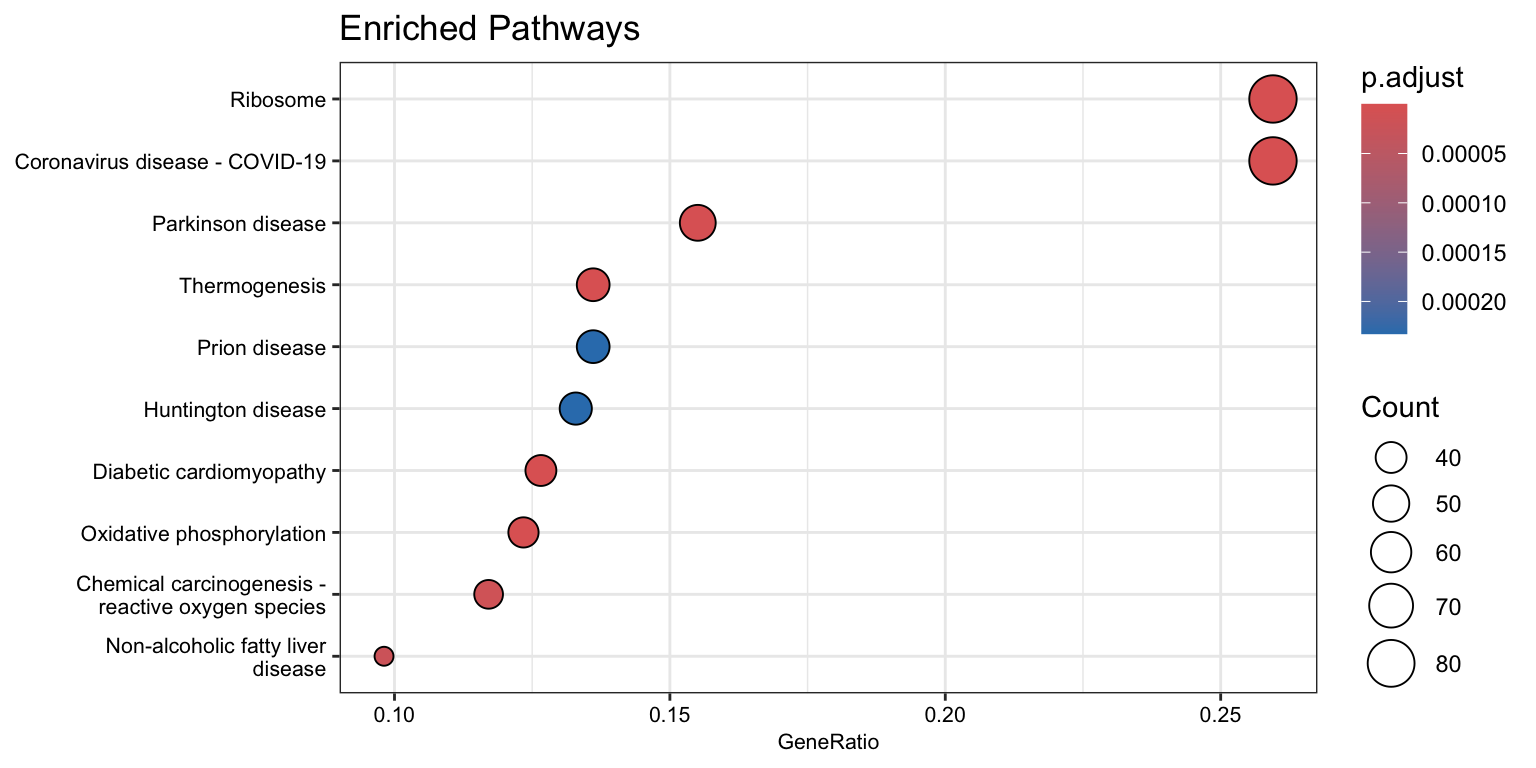
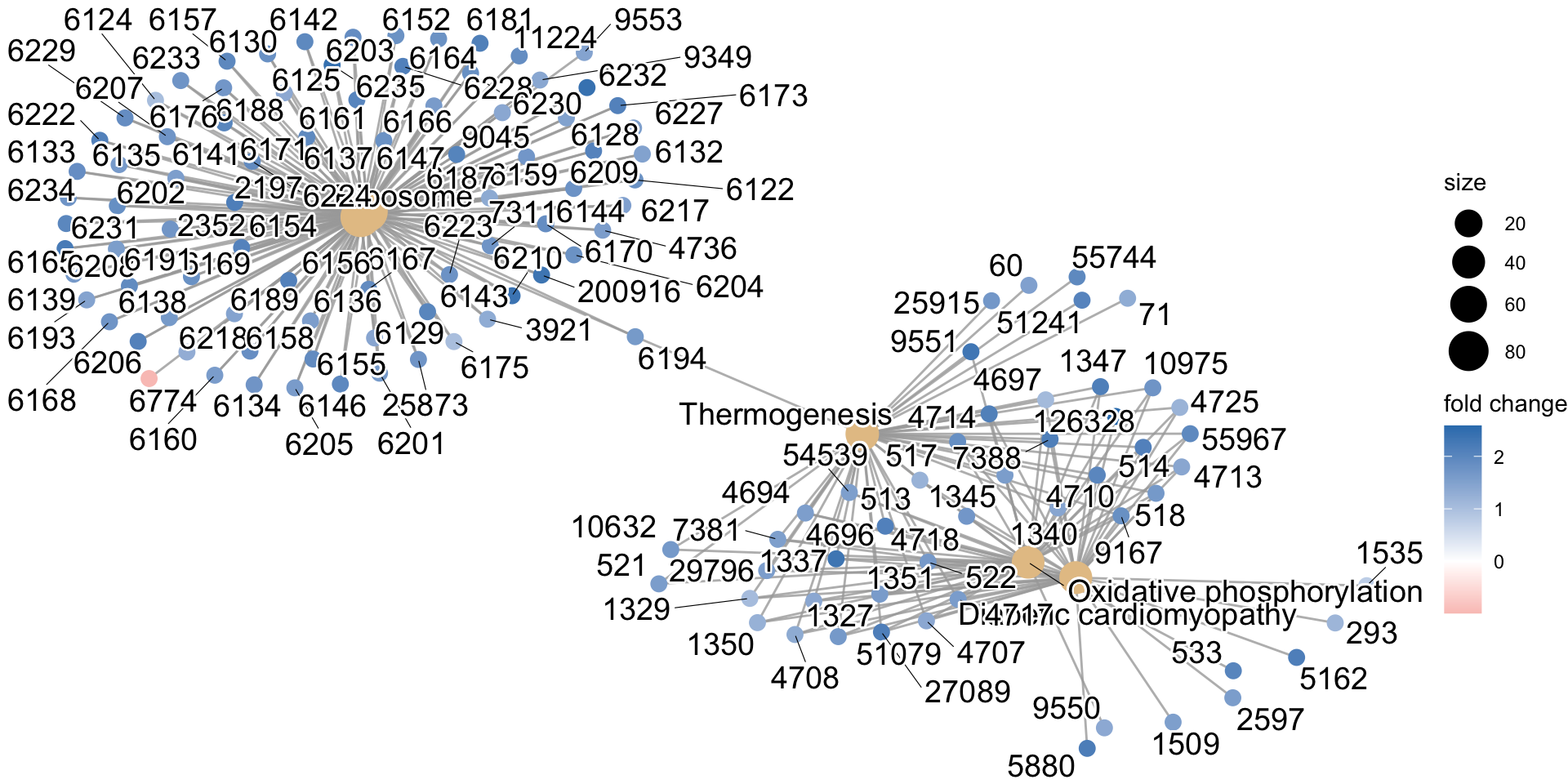
enrichKEGG object
R version 4.4.1 (2024-06-14)
Platform: x86_64-apple-darwin20
Running under: macOS Monterey 12.6
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/Chicago
tzcode source: internal
attached base packages:
[1] stats4 stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] UpSetR_1.4.0 SeuratObject_5.0.2
[3] sp_2.1-4 pathview_1.44.0
[5] org.Hs.eg.db_3.19.1 AnnotationDbi_1.66.0
[7] enrichplot_1.24.4 clusterProfiler_4.12.6
[9] reshape_0.8.9 gridExtra_2.3
[11] pheatmap_1.0.12 SingleCellExperiment_1.26.0
[13] SummarizedExperiment_1.34.0 Biobase_2.64.0
[15] GenomicRanges_1.56.2 GenomeInfoDb_1.40.1
[17] IRanges_2.38.1 S4Vectors_0.42.1
[19] BiocGenerics_0.50.0 MatrixGenerics_1.16.0
[21] matrixStats_1.4.1 ggpubr_0.6.0
[23] dplyr_1.1.4 ggplot2_3.5.1
loaded via a namespace (and not attached):
[1] splines_4.4.1 later_1.3.2 bitops_1.0-9
[4] ggplotify_0.1.2 tibble_3.2.1 R.oo_1.26.0
[7] polyclip_1.10-7 graph_1.82.0 XML_3.99-0.17
[10] lifecycle_1.0.4 httr2_1.0.5 rstatix_0.7.2
[13] rprojroot_2.0.4 globals_0.16.3 lattice_0.22-6
[16] MASS_7.3-61 backports_1.5.0 magrittr_2.0.3
[19] sass_0.4.9 rmarkdown_2.28 jquerylib_0.1.4
[22] yaml_2.3.10 httpuv_1.6.15 spam_2.11-0
[25] cowplot_1.1.3 DBI_1.2.3 RColorBrewer_1.1-3
[28] abind_1.4-8 zlibbioc_1.50.0 purrr_1.0.2
[31] R.utils_2.12.3 ggraph_2.2.1 RCurl_1.98-1.16
[34] yulab.utils_0.1.7 tweenr_2.0.3 rappdirs_0.3.3
[37] git2r_0.35.0 GenomeInfoDbData_1.2.12 ggrepel_0.9.6
[40] listenv_0.9.1 tidytree_0.4.6 parallelly_1.38.0
[43] codetools_0.2-20 DelayedArray_0.30.1 DOSE_3.30.5
[46] ggforce_0.4.2 tidyselect_1.2.1 aplot_0.2.3
[49] UCSC.utils_1.0.0 farver_2.1.2 viridis_0.6.5
[52] jsonlite_1.8.9 progressr_0.14.0 tidygraph_1.3.1
[55] Formula_1.2-5 ggnewscale_0.5.0 tools_4.4.1
[58] treeio_1.28.0 Rcpp_1.0.13 glue_1.8.0
[61] SparseArray_1.4.8 xfun_0.48 qvalue_2.36.0
[64] withr_3.0.1 fastmap_1.2.0 fansi_1.0.6
[67] digest_0.6.37 R6_2.5.1 gridGraphics_0.5-1
[70] colorspace_2.1-1 GO.db_3.19.1 RSQLite_2.3.7
[73] R.methodsS3_1.8.2 utf8_1.2.4 tidyr_1.3.1
[76] generics_0.1.3 data.table_1.16.2 graphlayouts_1.2.0
[79] httr_1.4.7 S4Arrays_1.4.1 scatterpie_0.2.4
[82] whisker_0.4.1 pkgconfig_2.0.3 gtable_0.3.5
[85] blob_1.2.4 workflowr_1.7.1 XVector_0.44.0
[88] shadowtext_0.1.4 htmltools_0.5.8.1 carData_3.0-5
[91] dotCall64_1.2 fgsea_1.30.0 scales_1.3.0
[94] png_0.1-8 ggfun_0.1.6 knitr_1.48
[97] rstudioapi_0.17.0 reshape2_1.4.4 nlme_3.1-166
[100] cachem_1.1.0 stringr_1.5.1 parallel_4.4.1
[103] pillar_1.9.0 grid_4.4.1 vctrs_0.6.5
[106] promises_1.3.0 car_3.1-3 Rgraphviz_2.48.0
[109] evaluate_1.0.1 KEGGgraph_1.64.0 cli_3.6.3
[112] compiler_4.4.1 rlang_1.1.4 crayon_1.5.3
[115] future.apply_1.11.2 ggsignif_0.6.4 labeling_0.4.3
[118] plyr_1.8.9 fs_1.6.4 stringi_1.8.4
[121] viridisLite_0.4.2 BiocParallel_1.38.0 munsell_0.5.1
[124] Biostrings_2.72.1 lazyeval_0.2.2 GOSemSim_2.30.2
[127] Matrix_1.7-1 patchwork_1.3.0 bit64_4.5.2
[130] future_1.34.0 KEGGREST_1.44.1 highr_0.11
[133] igraph_2.1.1 broom_1.0.7 memoise_2.0.1
[136] bslib_0.8.0 ggtree_3.12.0 fastmatch_1.1-4
[139] bit_4.5.0 ape_5.8 gson_0.1.0